Home \ Knowledge Hub \ Focus on \ Focus On… Foot and Mouth Disease – What’s in a strain


01 Jul 2015
Focus On… Foot and Mouth Disease – What’s in a strain
SHARE

ALASDAIR KING
Executive Director, International Veterinary Health
FMD virus is constantly evolving, creating a challenge for control programmes when considering vaccine strains. It is important to understand the circulating threats and to ensure that the vaccine strain being used is suitable for the region. Luckily, it is not necessary for a country to have a strain isolated from a current outbreak, or even from that specific country, in order to provide protection. It is usually enough to have a strain from the same lineage, or even sometimes from the same topotype.

The ever-changing virus
Outbreaks of FMD in the last year have led to questions being raised on the choice of strains being used in vaccines. FMD virus has continued to evolve with a complex variety of circulating strains which often do not provide cross-protection. As an example, O1 Manisa is no longer providing good matching in the laboratory to recent field strains. It is important to understand the spread of the virus and the relationship between serotypes, topotypes and lineages. Here we look at the strains and provide an explanation of the virus evolution.
As the FMD virus has continued to evolve and change over the years, some vaccine developers have managed to introduce new and better-matched vaccine strains. However, in order to make the best choice for a country, it is important to understand the strains and their relationships to each other.
FMD can be found in 7 different and distinct serotypes1 – O, A, C, Asia1, SAT1, SAT2, and SAT3. These serotypes are not distributed evenly around the world. For instance, the SAT (Southern African Territories) serotypes rarely spread outside of Africa. In addition, serotype C has not been identified anywhere since 2004 (note that it is unclear whether or not this means serotype C has been eradicated; there is some serological evidence that it may still be in circulation).
Serotypes
Serotypes O and A were first identified in 1922 and named after the areas where they were first found: Oise (in France) and Allemagne (Germany).2 The C serotype was reported in 1926. The SAT serotypes weren’t recognized until the late 1940’s, and finally, Asia1 was isolated in the early 1950’s. The key difference between all these serotypes is that they demonstrated no cross-protection against each other, and any immunity in cattle was limited to the individual serotype.
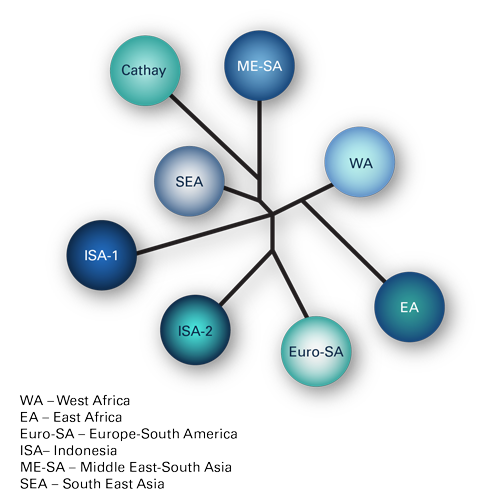
Topotypes, lineages and strains
The serotypes are further divided into topotypes, which are closely related groups of the virus, and these are generally geographically distinct. For instance, there are 8 topotypes for the O serotype, (see Figure 1). Then within the topotypes there may also be lineages which further subdivide the strains, (see Figure 2). Individual strains are usually named after the place in which they were isolated and the year. For instance, O/ME-SA/TUR/5/09 is the 5th strain of O serotype, ME-SA topotype to be isolated in Turkey during 2009 (the lineage is not included in the nomenclature). Some lineages have proven to be particularly aggressive and spread outside their expected geographic region to cause pandemics. For instance, between 1998 and 2001 the PanAsia lineage, (see Figure 3), extended from Asia to a number of normally FMD-free countries3, causing an explosion of outbreaks globally. Subsequent to this the PanAsia-2 lineage caused another pandemic. Additionally, while this is still circulating, there are now also concerns regarding the new lineage O/ME-SA/Ind-2001, which seems particularly aggressive and has appeared in a number of countries that had previously been free of disease for a significant period of time.
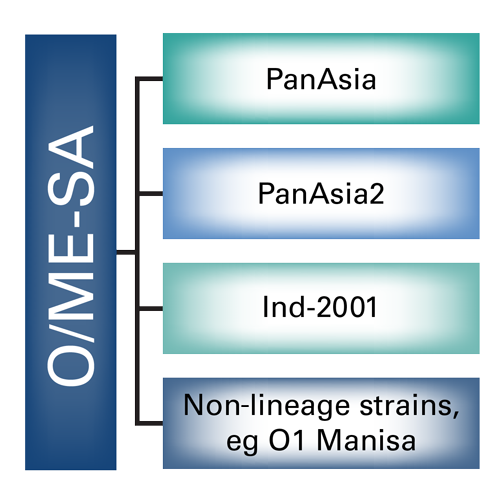
Protection
This variation means that one vaccine strain can never serve to protect the entire world. Countries and regions have very specific needs, and in order to handle this complexity the international FMD community has recognized seven pools to indicate which serotypes, topotypes and lineages are circulating (see Table 1). This list is kept up-to-date by the World Reference Laboratory for FMD at Pirbright, UK. Some countries on the edges of the pools may share viruses from more than one pool; for example, Egypt.1
Table 1 Circulating FMD Viruses
| Pool | Region | Serotype | Topotype | Lineage |
| 1 | Central/East Asia |
O O A (Asia1) |
SEA ME-SA ASIA |
Mya-98 PanAsia Sea-97 (Shamir) |
| 2 | South Asia |
O O A Asia1 |
ME-SA ME-SA ASIA ASIA |
PanAsia-2 Ind-2001 IND Sindh-08 |
| 3 | West Eurasia and Middle East |
O O A Asia1 |
ME-SA ME-SA ASIA ASIA |
PanAsia-2 Ind-2001 Iran-05 Sindh-08 |
| 4 | Eastern Africa |
O O O O A A SAT1 SAT2 |
ME-SA EA-2 EA-3 EA-4 ASIA AFRICA | Iran-05 |
| 5 | Western and Central Africa |
O A SAT1 SAT2 |
EA-3 AFRICA | |
| 6 | Southern Africa |
SAT1 SAT2 SAT3 (O) (A) | ||
| 7 | South America |
O A |
South Cone South Cone |
The more closely related a vaccine strain to the field strain, then the more probability of protection. There is usually good protection against field strains in the same lineage, and sometimes within the same topotype. Vaccine strains that can provide protection across topotypes are harder to find, and currently it is impossible to provide protection across serotypes. Therefore, when considering a vaccine strain choice, it is important to understand about the circulating risk.
Vaccine matching – the ongoing evolution
The reference vaccine strain ME-SA topotype O1/Manisa/TUR/69 is from a distinct single lineage3 that is markedly different from recent isolates. Vaccine matching reports from the WRLFMD, Pirbright, have shown that O1 Manisa is no longer matching well to the circulating strains. Continued use presents a real threat, and one that is unnecessary when more recent vaccine strains are available. We will cover vaccine matching further in a subsequent edition of this newsletter.
As the virus is constantly evolving, it is almost impossible to have a vaccine strain that has been developed from a current outbreak (it normally takes 6-18 months to develop new strains). However, by understanding the topotype and lineage of the outbreak, it is possible to select a vaccine strain that is closely related and therefore much more likely to provide protection. This is further enhanced by choosing high potency vaccines over 6PD50, as these generally provide better cross-protection.
When tendering for vaccine, authorities should consider the local threat and request companies to demonstrate the suitability of the offered vaccine strains.
References
1. Jamal SM, Belsham GJ. Foot-and-mouth disease: past, present and future. Vet Res. 2013;44:116.
2. Knowles, NJ. Council for National Academic Awards. Molecular and antigenic variation of foot and mouth disease virus. M Phil. Thesis. Available here. Accessed July 10, 2015.
3 Knowles NJ, Samuel AR, Davies PR, Midgley RJ, Valarcher JF. Pandemic strain of foot-and-mouth disease virus serotype O. Emerg Infect Dis. 2005;11(12):1887-1193.
ALASDAIR KING
Executive Director, International Veterinary Health

